Open Projects
Open Projects at the Bachelor, Master, Ph.D., and post doc level
We are always looking for talented and motivated students with backgrounds in physics, chemistry, biology, computer science and related disciplines who are interested in joining our lab for a range of projects. If you are interested, please contact Jan Lipfert with a CV, transcript, and short statement of motivation. Since the lab is moving to Utrecht University, all future projects will be at Utrecht in the Netherlands.
Highly parallized magnetic tweezers for single-molecule force spectroscopy
Master thesis project involving instrumentation, single-molecule measurements, and data analysis
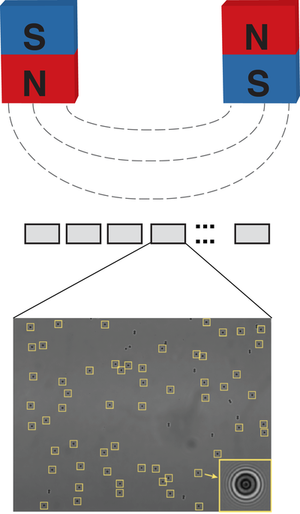
Magnetic tweezers are a powerful tool to apply forces and torques to single molecules tethered between a surface and small magnetic beads. A particular strength of magnetic tweezers is that we can apply constant forces to and track many molecules at the same time. Aim of this project is to develop a massively parallel magnetic tweezers set up that can probe 1000-10,000 molecules at the same time, with single molecule resolution. This new instrument will give us the possibility to probe ligand-receptor interactions and protein folding under forces at an unprecedented scale.
Deep Learning DNA-Protein Complexes
Master thesis project at the intersection of machine learning and biophysics
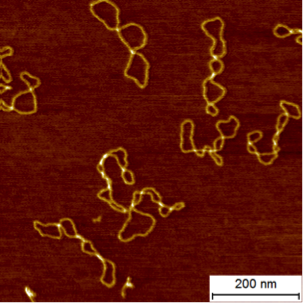
Atomic force microscopy (AFM) imaging is a very-high resolution type of scanning probe microscopy that can image biological macromolecules with a resolution of ~1 nm. AFM imaging has been used extensively to visualize DNA (an example is shown in the figure above), for example to probe its mechanical properties and to visualize its conformations. In addition, AFM imaging has proven to be powerful in revealing DNA-protein complexes involved a large range of biological process (see e.g. Vanderlinden et al., 2019).
In the past, AFM images of DNA and nucleo-protein complexes have mostly been analyzed manually, by clicking on or tracing structures in the images. Recently, we have developed algorithms that can process images automatically, by extracting and scoring relevant features based on machine learning techniques. Such automated analysis has the potential to 1) dramatically increase the throughput of AFM imaging analysis by replacing time consuming manual inspection (see Konrad et al. for automated analysis of 25,000 nucleosomes!) and 2) make the image analysis more reliable and statistically sound, by avoiding human judgment and biases.
A first application of the automated image analysis will be a full quantification of the topology of DNA plasmids (circular pieces of DNA present in bacteria; a typical image is shown above) to answer questions regarding the partitioning between DNA twist and writhe in these molecules (see Brouns et al. 2018). In a first step, we will transfer machine learning approaches based on convolutional neuronal networks that are used for vessel detection in medical image processing. If successful, the algorithmic approach can be extended to many other quantification tasks as well, for example to the analysis of the DNA-protein complexes or complex protein structures (see Müller et al., 2016 for an example).
We are looking for a highly motivated student from a background in physics, chemistry, or computer science who is interested in very interdisciplinary work and has basic programming experience in CPP and/or Matlab and/or Python. If you are interested, please contact me at Jan.Lipfert@lmu.de, including a brief CV and transcript.
References
- Konrad SF, Vanderlinden W, Lipfert J.
Quantifying epigenetic modulation of nucleosome breathing by high-throughput AFM imaging.
Biophys J. 2022 Mar 1;121(5):841-851. doi: 10.1016/j.bpj.2022.01.014. Epub 2022 Jan 20. - Brouns T., De Keersmaecker H., Konrad S.F., Kodera N., Ando T., Lipfert J., De Feyter S., Vanderlinden W. Free Energy Landscape and Dynamics of Supercoiled DNA by High-Speed Atomic Force Microscopy. ACS Nano. 2018 Oct 29. doi: 10.1021/acsnano.8b06994
- Müller, J.P., Mielke, S., Lof, A., Obser, T., Beer, C., Bruetzel, L.K., Pippig, D.A., Vanderlinden, W., Lipfert, J., Schneppenheim, R., et al. (2016). Force sensing by the vascular protein von Willebrand factor is tuned by a strong intermonomer interaction. Proc Natl Acad Sci U S A 113, 1208-1213.
- Vanderlinden W, Brouns T, Walker PU, Kolbeck PJ, Milles LF, Ott W, Nickels PC, Debyser Z, Lipfert J.
The free energy landscape of retroviral integration.
Nat Commun. 2019 Oct 18;10(1):4738. doi: 10.1038/s41467-019-12649-w

